You signed in with another tab or window. Reload to refresh your session.You signed out in another tab or window. Reload to refresh your session.You switched accounts on another tab or window. Reload to refresh your session.Dismiss alert
Copy file name to clipboardExpand all lines: lecture_notes/01_why_map.Rmd
+69-58Lines changed: 69 additions & 58 deletions
Original file line number
Diff line number
Diff line change
@@ -13,76 +13,79 @@ output:
13
13
toc: yes
14
14
---
15
15
16
-
# Setup
16
+
# Setup
17
17
18
18
```{r}
19
19
# Install packages
20
20
if (!require("pacman")) install.packages("pacman")
21
+
21
22
pacman::p_load(tidyverse, # tidyverse pkgs including purrr
22
23
tictoc, # performance test
23
24
broom, # tidy modeling
24
25
patchwork) # arranging ggplots
26
+
25
27
```
26
28
27
-
# Objectives
29
+
# Objectives
30
+
31
+
- How to use `purrr` to automate workflow in a cleaner, faster, and more extendable way
28
32
29
-
- How to use `purrr` to automate workflow in a cleaner, faster, and more extendable way
33
+
# Copy-and-paste programming
30
34
31
-
#Copy-and-paste programming
35
+
>Copy-and-paste programming, sometimes referred to as just pasting, is the production of highly repetitive computer programming code, as produced by copy and paste operations. It is primarily a pejorative term; those who use the term are often implying a lack of programming competence. It may also be the result of technology limitations (e.g., an insufficiently expressive development environment) as subroutines or libraries would normally be used instead. However, there are occasions when copy-and-paste programming is considered acceptable or necessary, such as for boilerplate, loop unrolling (when not supported automatically by the compiler), or certain programming idioms, and it is supported by some source code editors in the form of snippets. - Wikipedia
32
36
33
-
> Copy-and-paste programming, sometimes referred to as just pasting, is the production of highly repetitive computer programming code, as produced by copy and paste operations. It is primarily a pejorative term; those who use the term are often implying a lack of programming competence. It may also be the result of technology limitations (e.g., an insufficiently expressive development environment) as subroutines or libraries would normally be used instead. However, there are occasions when copy-and-paste programming is considered acceptable or necessary, such as for boilerplate, loop unrolling (when not supported automatically by the compiler), or certain programming idioms, and it is supported by some source code editors in the form of snippets. - Wikipedia
37
+
- The following exercise was inspired by [Wickham's example](http://adv-r.had.co.nz/Functional-programming.html).
34
38
35
-
-The following exercise was inspired by [Wickham's example](http://adv-r.had.co.nz/Functional-programming.html).
39
+
- Let's imagine `df` is a survey data.
36
40
37
-
- Let's imagine `df` is a survey data.
41
+
- a, b, c, d = Survey questions
38
42
39
-
- a, b, c, d = Survey respondents
43
+
- -99: non-responses
44
+
45
+
- Your goal: replace -99 with NA
40
46
41
-
- -99: non-responses
42
-
43
-
- Your goal: replace -99 with NA
44
-
45
47
```{r}
46
48
# Data
47
49
df <- tibble("a" = -99,
48
50
"b" = -99,
49
51
"c" = -99,
50
52
"d" = -99)
51
-
53
+
54
+
```
55
+
56
+
57
+
```{r}
52
58
# Copy and paste
53
59
df$a[df$a == -99] <- NA
54
60
df$b[df$b == -99] <- NA
55
61
df$c[df$c == -99] <- NA
56
62
df$d[df$d == -99] <- NA
57
63
58
-
df
59
-
60
64
```
61
65
62
-
-**Challenge 1**. Explain why this solution is not very efficient. (e.g., If `df$a[df$a == -99] <- NA` has an error, how are you going to fix it?) A solution is not scalable if it's not automatable and, thus, scalable.
66
+
-**Challenge 1**. Explain why this solution is not very efficient. (e.g., If `df$a[df$a == -99] <- NA` has an error, how are you going to fix it?) A solution is not scalable if it's not automatable and, thus, scalable.
63
67
64
-
# Using a function
68
+
# Using a function
65
69
66
-
- Let's recall what's function in R: input + computation + output
70
+
- Let's recall what's function in R: input + computation + output
67
71
68
-
- If you write a function, you gain efficiency because you don't need to copy and paste the computation part.
72
+
- If you write a function, you gain efficiency because you don't need to copy and paste the computation part.
69
73
70
-
`
71
-
function(input){
72
-
73
-
computation
74
-
75
-
return(output)
74
+
\` function(input){
76
75
77
-
}
78
-
`
76
+
computation
77
+
78
+
return(output)
79
+
80
+
} \`
79
81
80
82
```{r}
81
83
82
84
# Function
83
85
fix_missing <- function(x) {
84
86
x[x == -99] <- NA
85
-
x
87
+
# This is better
88
+
return(x)
86
89
}
87
90
88
91
# Apply function to each column (vector)
@@ -91,34 +94,30 @@ df$b <- fix_missing(df$b)
91
94
df$c <- fix_missing(df$c)
92
95
df$d <- fix_missing(df$d)
93
96
94
-
df
95
-
96
97
```
97
98
98
-
-**Challenge 2** Why using function is more efficient than 100% copying and pasting? Can you think about a way we can automate the process?
99
+
-**Challenge 2** Why using function is more efficient than 100% copying and pasting? Can you think about a way we can automate the process?
99
100
100
-
- Many options for automation in R: `for loop`, `apply` family, etc.
101
+
- Many options for automation in R: `for loop`, `apply` family, etc.
101
102
102
-
- Here's a tidy solution comes from `purrr` package.
103
+
- Here's a tidy solution comes from `purrr` package.
103
104
104
-
- The power and joy of one-liner.
105
+
- The power and joy of one-liner.
105
106
106
107
```{r}
107
108
108
-
df <- purrr::map_df(df, fix_missing)
109
-
110
-
df
109
+
purrr::map_df(df[,column], fix_missing)
111
110
112
111
```
113
112
114
-
`map()` is a [higher-order function](https://en.wikipedia.org/wiki/Map_(higher-order_function)) that applies a given function to each element of a list/vector.
113
+
`map()` is a [higher-order function](https://en.wikipedia.org/wiki/Map_(higher-order_function)) that applies a given function to each element of a list/vector.
115
114
116
115
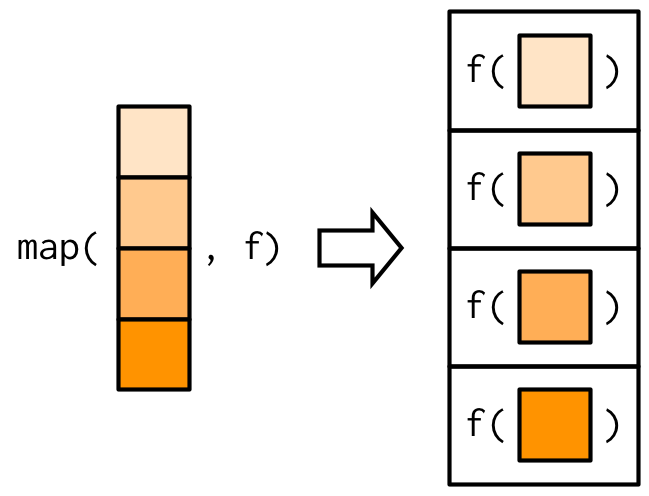
117
116
118
117
- Input: Takes a vector/list.
119
-
118
+
120
119
- Computation: Calls the function once for each element of the vector
121
-
120
+
122
121
- Output: Returns in a list or whatever data format you prefer (e.g., `_df helper: dataframe`)
123
122
124
123
**Challenge 3** If you run the code below, what's going to be the data type of the output?
@@ -128,13 +127,16 @@ df
128
127
map_chr(df, fix_missing)
129
128
130
129
```
131
-
- Why `map()` is a good alternative to `for loop`. (For more information, watch Hadley Wickam's talk titled ["The Joy of Functional Programming (for Data Science)"](https://www.youtube.com/watch?v=bzUmK0Y07ck&ab_channel=AssociationforComputingMachinery%28ACM%29).)
132
130
133
-
```{r}
131
+
- Why `map()` is a good alternative to `for loop`. (For more information, watch Hadley Wickam's talk titled ["The Joy of Functional Programming (for Data Science)"](https://www.youtube.com/watch?v=bzUmK0Y07ck&ab_channel=AssociationforComputingMachinery%28ACM%29).)
134
132
133
+
```{r}
135
134
# Built-in data
136
135
data("airquality")
137
136
137
+
```
138
+
139
+
```{r}
138
140
# 0.029 sec elapsed
139
141
tic()
140
142
@@ -147,8 +149,11 @@ for (i in seq_along(airquality)) { # Sequence variable
- In short, `map()` is more readable, faster, and easily extensive with other data science tasks (e.g., wrangling, modeling, and visualization) using `%>%`.
161
165
162
-
- Final point: Why not base R `apply` family?
166
+
- In short, `map()` is more readable, faster, and easily extensive with other data science tasks (e.g., wrangling, modeling, and visualization) using `%>%`.
167
+
168
+
- Final point: Why not base R `apply` family?
163
169
164
-
Short answer: `purrr::map()` is simpler to write. For instance,
170
+
Short answer: `purrr::map()` is simpler to write. For instance,
Performance testing (profiling) is an important part of programming. `tictic()` measures the time that needs to take to run a target function for once. If you want a more robust measure of timing as well as information on memory (**speed** and **space** both matter for performance testing), consider using the [`bench` package](https://github.com/r-lib/bench) that is designed for high precising timing of R expressions.
171
-
176
+
Performance testing (profiling) is an important part of programming. `tictic()` measures the time that needs to take to run a target function for once. If you want a more robust measure of timing as well as information on memory (**speed** and **space** both matter for performance testing), consider using the [`bench` package](https://github.com/r-lib/bench) that is designed for high precising timing of R expressions.
172
177
173
178
```{r}
174
179
map_mark <- bench::mark(
@@ -180,11 +185,11 @@ map_mark <- bench::mark(
180
185
map_mark
181
186
```
182
187
183
-
# Applications
188
+
# Applications
184
189
185
-
1. Many models
190
+
1. Many models
186
191
187
-
- One popular application of `map()` is to run regression models (or whatever model you want to run) on list-columns. No more copying and pasting for running many regression models on subgroups!
192
+
- One popular application of `map()` is to run regression models (or whatever model you want to run) on list-columns. No more copying and pasting for running many regression models on subgroups!
- For more information on this technique, read the Many Models subchapter of the [R for Data Science](https://r4ds.had.co.nz/many-models.html#creating-list-columns).
203
+
- For more information on this technique, read the Many Models subchapter of the [R for Data Science](https://r4ds.had.co.nz/many-models.html#creating-list-columns).
@@ -79,6 +79,7 @@ for (univ in c("Berkeley", "Stanford")) {
79
79
}
80
80
81
81
```
82
+
82
83
- This is not bad, but ... n arguments -> n nested for loops. As a scale of your problem grows, your code gets really complicated.
83
84
84
85
> To become significantly more reliable, code must become more transparent. In particular, nested conditions and loops must be viewed with great suspicion. Complicated control flows confuse programmers. Messy code often hides bugs. — [Bjarne Stroustrup](https://en.wikipedia.org/wiki/Bjarne_Stroustrup)
@@ -90,30 +91,37 @@ for (univ in c("Berkeley", "Stanford")) {
90
91
**Challenge 2** Why are we using `rep()` to create input vectors? For instance, for `univ_list` why not just use `c("Berkeley", "Stanford")`?
91
92
92
93
```{r}
93
-
94
94
# Inputs (remember the length of these inputs should be identical)
0 commit comments