You signed in with another tab or window. Reload to refresh your session.You signed out in another tab or window. Reload to refresh your session.You switched accounts on another tab or window. Reload to refresh your session.Dismiss alert
@@ -253,16 +253,6 @@ Get started with the genetic algorithm by reading the tutorial titled [**Introdu
253
253
254
254
[](https://www.linkedin.com/pulse/introduction-optimization-genetic-algorithm-ahmed-gad)
255
255
256
-
## Tutorial: Build Neural Networks in Python
257
-
258
-
Read about building neural networks in Python through the tutorial titled [**Artificial Neural Network Implementation using NumPy and Classification of the Fruits360 Image Dataset**](https://www.linkedin.com/pulse/artificial-neural-network-implementation-using-numpy-fruits360-gad) available at these links:
*[Towards Data Science](https://towardsdatascience.com/artificial-neural-network-implementation-using-numpy-and-classification-of-the-fruits360-image-3c56affa4491)
## Tutorial: Optimize Neural Networks with Genetic Algorithm
267
257
268
258
Read about training neural networks using the genetic algorithm through the tutorial titled [**Artificial Neural Networks Optimization using Genetic Algorithm with Python**](https://www.linkedin.com/pulse/artificial-neural-networks-optimization-using-genetic-ahmed-gad) available at these links:
@@ -273,29 +263,6 @@ Read about training neural networks using the genetic algorithm through the tuto
273
263
274
264
[](https://www.linkedin.com/pulse/artificial-neural-networks-optimization-using-genetic-ahmed-gad)
275
265
276
-
## Tutorial: Building CNN in Python
277
-
278
-
To start with coding the genetic algorithm, you can check the tutorial titled [**Building Convolutional Neural Network using NumPy from Scratch**](https://www.linkedin.com/pulse/building-convolutional-neural-network-using-numpy-from-ahmed-gad) available at these links:
[This tutorial](https://www.linkedin.com/pulse/building-convolutional-neural-network-using-numpy-from-ahmed-gad)) is prepared based on a previous version of the project but it still a good resource to start with coding CNNs.
286
-
287
-
[](https://www.linkedin.com/pulse/building-convolutional-neural-network-using-numpy-from-ahmed-gad)
288
-
289
-
## Tutorial: Derivation of CNN from FCNN
290
-
291
-
Get started with the genetic algorithm by reading the tutorial titled [**Derivation of Convolutional Neural Network from Fully Connected Network Step-By-Step**](https://www.linkedin.com/pulse/derivation-convolutional-neural-network-from-fully-connected-gad) which is available at these links:
*[Towards Data Science](https://towardsdatascience.com/derivation-of-convolutional-neural-network-from-fully-connected-network-step-by-step-b42ebafa5275)
[](https://www.linkedin.com/pulse/derivation-convolutional-neural-network-from-fully-connected-gad)
298
-
299
266
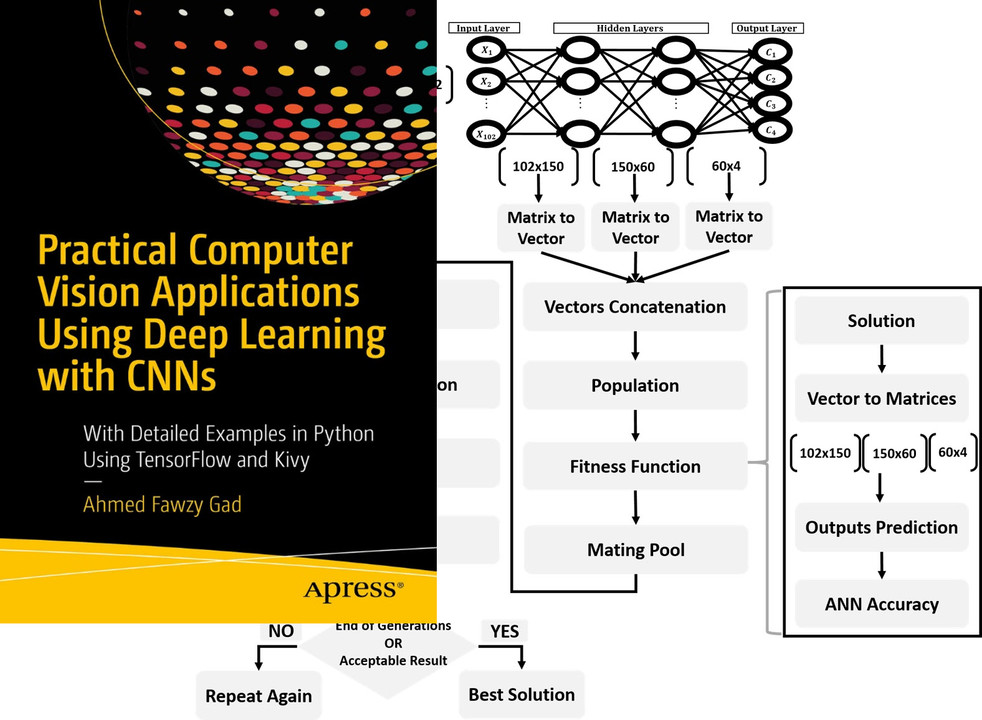
## Book: Practical Computer Vision Applications Using Deep Learning with CNNs
300
267
301
268
You can also check my book cited as [**Ahmed Fawzy Gad 'Practical Computer Vision Applications Using Deep Learning with CNNs'. Dec. 2018, Apress, 978-1-4842-4167-7**](https://www.amazon.com/Practical-Computer-Vision-Applications-Learning/dp/1484241665) which discusses neural networks, convolutional neural networks, deep learning, genetic algorithm, and more.
raiseException("The output from the gene_constraint callable/function must be a list or NumPy array that is subset of the passed values (second argument).")
158
175
159
176
# After going through all the values, check if any value satisfies the constraint.
160
-
iflen(filtered_values_indices) >0:
177
+
iflen(filtered_values) >0:
161
178
# At least one value was found that meets the gene constraint.
162
179
pass
163
180
else:
164
181
# No value found for the current gene that satisfies the constraint.
165
-
ifnotself.suppress_warnings:
166
-
warnings.warn(f"No value found for the gene at index {gene_idx} with value {solution[gene_idx]} that satisfies its gene constraint.")
182
+
ifnotself.suppress_warnings: warnings.warn(f"Failed to find a value that satisfies its gene constraint for the gene at index {gene_idx} with value {solution[gene_idx]} at generation {self.generations_completed}.")
ifnotself.suppress_warnings: warnings.warn(f"Failed to find a unique value for gene with index {duplicate_index} whose value is {solution[duplicate_index]}. Consider adding more values in the gene space or use a wider range for initial population or random mutation.")
72
+
ifnotself.suppress_warnings: warnings.warn(f"Failed to find a unique value for gene with index {duplicate_index} whose value is {solution[duplicate_index]} at generation {self.generations_completed}. Consider adding more values in the gene space or use a wider range for initial population or random mutation.")
ifnotself.suppress_warnings: warnings.warn(f"Failed to find a unique value for gene with index {duplicate_index} whose value is {solution[duplicate_index]}. Consider adding more values in the gene space or use a wider range for initial population or random mutation.")
323
+
ifnotself.suppress_warnings: warnings.warn(f"Failed to find a unique value for gene with index {duplicate_index} whose value is {solution[duplicate_index]} at generation {self.generations_completed+1}. Consider adding more values in the gene space or use a wider range for initial population or random mutation.")
0 commit comments